Genetic Diversity of Tomato Yellow Leaf Curl Virus (TYLCV) in Mauritius
| Received 06 Feb, 2024 |
Accepted 06 May, 2024 |
Published 07 May, 2024 |
Background and Objective: Tomato yellow leaf curl disease (TYLCD), caused by tomato yellow leaf curl virus (TYLCV), was first reported in Mauritius in 2009. This study reports on the status regarding the genetic identity of TYLCV isolates in Mauritius, based on data collected in 2015-2018. Materials and Methods: The TYLCV-infected plants showing clear symptoms were collected from 21 localities around the island. The presence of TYLCV was confirmed using standard TAS-ELISA tests and triplicates of each leaf sample were subject to DNA extraction followed by PCR amplification of part of the C1 replication-associated gene and the PCR products were sequenced and edited and subjected to a similarity search using the BLASTN tool. The sequences were then aligned using MEGA11 software and phylogenetic trees were generated using the UPGMA method with 1000 bootstrap values. Results: This study revealed 12 genetically distinct isolates of TYLCV, with 4 of them seemingly representing the ancestral TYLCV-IL lineage, 6 of them representing variants or recombinants of the ancestral lineage and 2 isolates which could potentially represent new strains. This study shows that, over a period of 9 years, with the continued cultivation of susceptible tomato varieties and the presence of alternate hosts, the original 2009 TYLCV isolate underwent multiple mutation events that eventually resulted in the appearance of two possibly novel strains. Conclusion: The TYLCV has shown a very rapid rate of mutation on Mauritius Island, possibly evolving into new strains and now constitutes an important economic threat to local tomato production.
INTRODUCTION
In Mauritius, tomato is one of the most important cultivated and consumed vegetable crops, with approximately 14,000 ton produced each year and contributes substantially to food security on the island1. However, tomato production on the island is erratic, varying between 8000-11,000 ton per year, as fruit yield is subject to biotic and abiotic constraints. Abiotic factors affecting yield in Mauritius include rainfall, temperature extremes and poor soil conditions while biotic factors comprise mostly of major tomato diseases including tomato yellow leaf curl disease (TYLCD). The TYLCD is caused by the species tomato yellow leaf curl virus (TYLCV) which affects tomato harvests across warm and temperate climates all around the globe inducing up to 100% yield loss, specifically in susceptible varieties and is transmitted by a single insect species, the whitefly Bemisia tabaci. Besides tomato (Solanum lycopersicum) which serves as the primary host, this icosahedral Begomovirus infects over 12 plant families2, mostly dicots. Belonging to the plant-infecting DNA Geminiviridae family3, there are various TYLCV species and strains, collectively termed the tomato yellow leaf curl virus-like viruses4. The first predominantly whitefly-vectored TYLCVs originated in the Jordan Valley, Israel, in the 1930s5 and it was not until the 1960s that TYLCV was officially identified as the causal virus of this disease6. Lefeuvre et al.7, report that the Israel strain (TYLCV-IL) and Mild strain (TYLCV-Mld), from the Middle East, form two major ancestral clades from where other recombinant strains have arisen and spread from and across the Mediterranean islands/countries, even reaching subtropical/tropical south oceanic islands. Currently, there are thirteen virus species causing tomato yellow leaf curl disease (TYLCD) according to the International Committee on Taxonomy of Viruses list of 20196. These viruses are grouped into monopartite and bipartite viruses based on the number of DNA genome components. The complete names of the viruses are tomato yellow leaf curl (TYLC) Axarquia virus (TYLCAxV-[ES]), TYLC-Guangdong virus (TYLCGuV-[CN]), TYLC-Indonesia virus (TYLCIDV-[Lem]), TYLC-Malaga virus (TYLCMaV-[ES]), TYLC-Mali virus (TYLCMLV-[ML]), TYLC-Sardinia virus (TYLCSV-[IT]), TYLC-Shuangbai virus (TYLCShV-[CN]), TYLC-Vietnam virus (TYLCVNV-[VN]), TYLC-China virus (TYLCCNV-[CN]), TYLC-Yunnan virus (TYLCYnV-[CN]), TYLC-Kanchanaburi virus (TYLCKaV-[VN]) and TYLC-Thailand virus (TYLCTHV-[MM]).
The TYLCV was first reported in Mauritius in 2009, after TYLCD symptoms (such as leaf curling, vein yellowing and dwarfing) and whitefly populations were observed in tomato open fields, mainly on tomato varieties in the southern regions of the island by Lobin et al.8. Authors report that sequence analysis of this TYLCV isolate collected in 2009 showed that it had 100% identity with nucleotides 458 to 1,036 of an isolate from Almeria (Spain) (GenBank Accession no. AJ489258) and isolates from the Netherlands (FJ439569), Morocco (EF060196) and Spain (AJ519441) and nucleotides 451 to 1,029 of the RE4 isolate from Reunion Island (AM409201).
For the genetic characterization of TYLCV strains worldwide, there are a number of protocols and primers which have been used. The TY1 and TY2 primers were used by Accotto et al.9 to amplify a DNA fragment of about 580 bp in the gene encoding the coat protein (CP) of TYLCD-associated viruses present in the Mediterranean basin. They successfully amplified this gene fragment from tomato yellow leaf curl-Sardinia virus (TYLCV-SV) and tomato yellow leaf curl-Israel virus (TYLCV-IL) and this primer pair has since been used in various countries to screen against TYLCD10,11. The C2-specific primer pairs MA13 and MA17, amplify a 348 bp fragment corresponding to 85% of the C2 open reading frame. The C2 gene was selected to design wide range primers for PCR because it contains regions that are well conserved among monopartite TYLCV species9. Primers TYLCV-1840F/IL-2642R (targeting the C1 gene) and TYLCV-1840F/ Mld-2354R were developed in order to monitor the rapidly expanding worldwide TYLCD epidemics and were used in a multiplex polymerase chain reaction assay to detect and characterize the TYLCV-IL and TYLCV-Mld clade isolates7. Primer pair TY255/TY2463 was designed to screen TYLCV and tomato yellow leaf curl sardinia virus (TYLC-SV) and recombinants between these, such as tomato yellow leaf curl malaga virus (TYLC-MalV) generating DNA fragments of specific sizes for each virus species and recombinant: 800 bp for TYLC-SV, 410 bp for TYLCV and 570 bp for TYLC-MalV12.
In Mauritius, since 2009, there was no further molecular analysis of local TYLCV isolates and concern about the virus and its associated disease lapsed until 2015-2020, when a resurgence of TYLCV was observed in local tomato fields across the island. Given the rapid rate of evolution of phytopathogens like TYLCV via recombination, reassortment and mutation, genetic variation can effectively engender novel TYLCV variants or strains with enhanced ecological adaptation across a relatively short time span, facilitating their spread and epidemiology. Sequence information generated in this study thus allowed an investigation of not only the extent of relatedness, or genetic diversity, among our local isolates but also a comparison between the local and international TYLCV isolates obtained from various locations around the globe. With TYLCV dissemination aggravating agro-harvesting systems and contributing to food insecurity, coupled with problems resulting from international trade, this study provides an improved insight into how TYLCV diversity has evolved in Mauritius over a period of 9 years. This would aid in improving the design of phytosanitary measures and field infection management, while adjusting trade or breeding policy to prevent and/or reduce potential outbreaks and propagation.
MATERIALS AND METHODS
Sampling and serological testing: Sampling was carried out from November, 2015 to August, 2018 in open fields and greenhouses under tomato cultivation over 21 localities across Mauritius Island. Sampling was carried out based on symptoms observation including leaf curl, leaf yellowing, stunting and poor fruit set. Leaves which were sampled were then subjected to serological analysis through TAS-ELISA using a commercial kit (ADGEN Phytodiagnostics, Neogen Europe LTD, Scotland, UK). Procedures for detection of the virus were based on Manufacturer’s protocol. Leaves which tested positive were then kept at -80°C for molecular analysis.
DNA extraction, PCR and sequencing: The TYLCV isolates were collected on the island between November, 2015 and August, 2018 on symptomatic tomato leaves from the varieties Jessica, Typhoon, Swaraksha and Rose, across 21 localities covering the whole island and which tested positive with TYLCV-specific TAS-ELISA tests. DNA extraction was performed on 3 replicates of the 21 leaf samples using a DNeasy® Plant Mini Kit (Qiagen) following the manufacturer’s instructions. The DNA yield and purity were determined using standard spectrophotometry at 260 and 280 nm. Out of the 21 leaf samples, only 14 yielded DNA of sufficient quantity and quality to proceed to PCR. The positive results for these 14 samples tested were obtained with primers pairs MA17/MA13, TY255/TY2463 and TYLCV-1840F/IL-2642R.
The PCR was performed using the primer pair TYLCV-1840F/IL-2642R, which codes for the TYLCV C1 replication-associated gene, in a 25 μL reaction mixture containing 1.25 mM MgCl2, 1×PCR buffer, 0.2 μM of each dNTP, 2 μM of each primer and 1.0 U of GoTaqp® DNA polymerase (Promega) in a GeneAmp PCR System 9600 thermocycler (Perkin-Elmer, USA). The PCR thermal profile was: Pre-PCR denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1.5 min and extension at 72°C for 1 min and a final extension at 72°C for 5 min. Standard gel electrophoresis with 10 μL of the final reaction mixture on a 1.5% agarose gel was used to separate PCR products. Ethidium bromide stained DNA fragments were visualised on agarose gels using ultraviolet fluorescence. The PCR amplicons of the correct size were purified using a Wizard® SV Gel and PCR Clean-Up System (Promega) following the manufacturer's instructions. Sequencing of the amplified PCR products was carried out using the dideoxy (Sanger) method of sequencing.
Sequence analysis and phylogeny: The DNA sequence chromatograms were viewed and edited using the software tool FinchTV Version 1.4 (Copyright Geospiza Inc). The edited sequences were then subjected to a similarity search using the BLASTN tool from NCBI to find similar corresponding matches. The sequences were then aligned using MEGA version 11 (ClustalW alignment option) and phylogenetic trees were generated using the UPGMA method with 1000 bootstrap values.
RESULTS
PCR results and analysis of sequences: Amplification of extracted DNA samples using the primer pair TYLCV-1840F/IL-2642R produced the expected 802 bp PCR product (Fig. 1). The correctly-sized bands were excised from the agarose gel, purified and the purification products were checked again on agarose to confirm their presence before being sent for sequencing. The BLAST analysis of the sequences showed that all 14 isolates matched with >99% sequence similarity to two previously collected TYLCV isolates from the database, namely a TYLCV isolate collected from France (Accession number MG489967.1) and a TYLCV isolate previously collected in Mauritius in 2009 (Accession number KX347172.1). A pairwise sequence alignment of these two sequences showed that they were genetically identical at this locus, raising the possibility that these might actually be the same TYLCV strain. It seemed therefore apparent that all the isolates collected in Mauritius in the period 2015-2018 were genetic variants of this ancestral strain, which existed on the island since 2009. Sequence alignment of the 14 sequences collected from Mauritius showed that three of these isolates were genetically identical, therefore reducing the number of genetically distinct isolates to 12, which were then named Isolates A, B, C, D, E, F, G, H, I, J, K and L (referred to as Seq A-L in Fig. 2 and 3). The sequences were submitted to GenBank and published with accession numbers MW691996-MW692007.
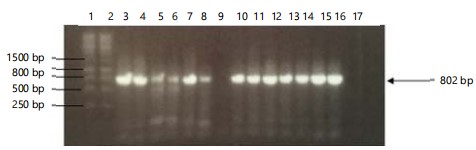
|
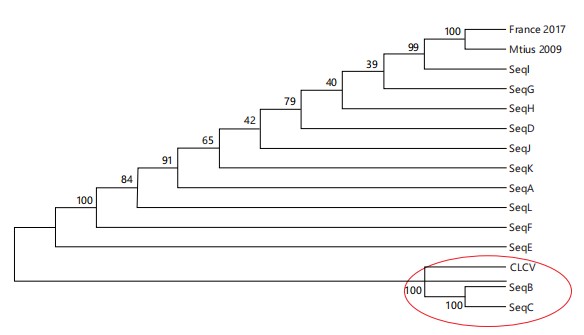
|
Phylogenetic analysis of mauritian isolates: In order to investigate the genetic relatedness of the locally collected TYLCV isolates, a phylogenetic analysis was performed with all 12 isolates and an outgroup virus, the Chili leaf curl virus (ChiLCV), which was picked up in the BLAST similarity search results (Fig. 2). This shows that all the 12 collected Mauritian isolates are genetic variants of the single ancestral isolate which was present on the island since 2009, however, isolates B and C exhibit a great genetic distance from the rest of the isolates, being more closely related to the outgroup virus, ChiLCV, than to all the other isolates. The clade containing Isolates B and C and ChiLCV branches off separately from all the other clades very early in the phylogenetic tree, clearly indicating that these two TYLCV isolates are genetically distinct from the other ones.
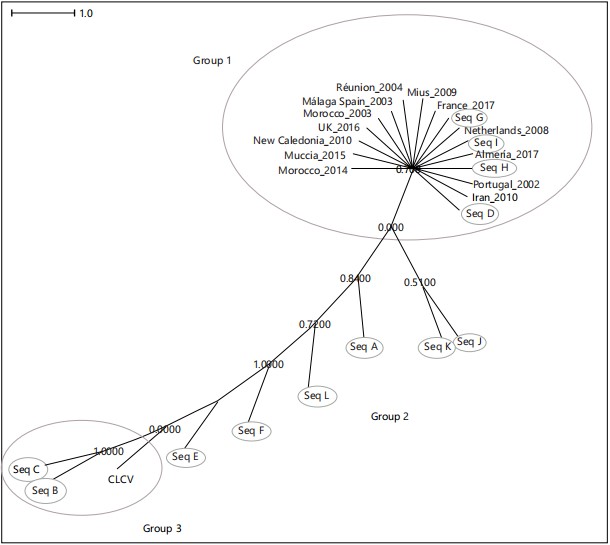
|
Phylogenetic analysis of mauritian isolates with international TYLCV isolates: From the BLAST similarity search results, it was possible to pick out TYLCV sequences from 13 other countries in order to compare them with the Mauritian isolates. This is shown in Fig. 3. This tree very clearly shows that most of the international isolates are genetic variants, or descend from, a single ancestral lineage that spread across Europe (UK, Spain, France, Netherlands, Portugal), the Middle East (Iran), Northern Africa (Morocco), the South Pacific (New Caledonia) and the Indian Ocean (Mauritius, Réunion Island), indicating a very widespread distribution of this ancestral lineage representing most probably the Israel strain (TYLCV-IL) and/or the mild strain (TYLCV-Mld). However, surprisingly, isolates B and C recently collected in Mauritius, cluster very far away from this ancestral lineage, raising the possibility that they could represent a new TYLCV strain.
DISCUSSION
The detailed phylogenetic analyses done in this study indicate that the twelve isolates collected from Mauritius from 2015-2018 can be roughly classified into three “groups” (Fig 3). Group 1 represents the “old” 2009 lineage, a recombinant between TYLCV-IL and TYLCSV-ES, which is known as the severe strain and which is widely distributed around the world and Isolates G, I, H and D fall in this group. Group 2 represents other recombinants derived from TYLCV-IL which have probably evolved through mutation of the old 2009 lineage; this group represents Isolates E, F, L, A, K, J. Group 3 comprises of Isolate B and C and these show a much higher degree of genetic polymorphism from the rest of the isolates and, although they too are derived from the parental TYLCV-IL, these two isolates could represent new strains. It is known that the C1 gene is one of the principal recombination hotspots in the TYLCV genome, besides the C4 region and the non-coding IR (Intergenic Region)6 and the single nucleotide polymorphisms (SNPs) in this gene, as well as other genes, accumulated over a period of 9 years have possibly led to the emergence of new strains of TYLCV with enhanced biological capabilities on the island.
The possibility of Isolates B and C belonging to a new strain of the virus is further supported by the fact that these isolates cluster closer to an outgroup virus, the Chili leaf curl virus (ChiLCV), than to the ancestral TYLCV-IL lineage. New viral strains emerge as a result of mutation, inter-genomic recombination13 as well as genetic reassortment of strains belonging to the same or related species, a common evolutionary mechanism in both RNA and DNA viruses. Mixed infections also provide an important platform to favor mutation through recombination and a high degree of intra- and inter-species recombination has been observed within the TYLCV complex or among begomoviruses during mixed infections6. For example, TYLCV-IL is the result of recombination between TYLCV-Mld and tomato leaf curl Karnataka virus (ToLCKV, a tomato-infecting begomovirus) which occurred in nature during mixed infection14. Likewise, the Sardinia strain (TYLCSV) most probably emerged from the South African cassava mosaic virus (SACMV) via genetic recombination4 and tomato yellow leaf curl Málaga virus (TYLCMaV) and tomato yellow leaf curl Axarquia virus (TYLCAxV) are also thought to have emerged from co-infection of tomato plants with TYLCV and TYLCSV and these recombinant viruses display a broader host range than either of the parents15. Since it is not known whether other TYLCV-like viruses may also have been introduced to Mauritius, the possibility of Isolates B and C being recombinants cannot be ruled out. Interestingly, BLAST analysis of the sequences derived from the amplification of part of the C2 gene using the primer set MA13/MA17 sequenced in this study show that Isolates B and C show a 98% match to the African cassava mosaic virus (ACMV), thus raising the possibility that Isolates B and C are recombinants of TYLCV-IL and ACMV. Although, there are no published reports by local researchers of the presence of ACMV in Mauritius, it is listed as being present in Mauritius in the CABI compendium by Rey et al.16.
The genetic identity and diversity of the TYLCV strains in Mauritius provide us with important clues regarding their origin and migration paths. The Mediterranean Basin (specifically Israel) has been the source of several TYLCV strains, variants and recombinants, which have then travelled worldwide. Our phylogenetic tree presented in Fig. 3 clearly shows that TYLCV isolates collected from 10 countries spanning the European continent, Western Asia, South Africa, the Pacific (New Caledonia) and the Indian Ocean are likely to represent a single clonal lineage, the “ancestral” TYLCV-IL lineage mentioned by Lefeuvre et al.7, which quickly spread worldwide and underwent various recombination events. The Mauritian TYLCV isolate collected and analyzed in 2009 was itself a recombinant between a TYLCV-IL strain collected from Reunion Island in 2004 (Accession No AM409201) and the Spanish strain of TYLCSV collected from Morocco (Agadir) in 2012, named TYLCSV-ES (Accession No LN846598). It can be speculated that this Mauritian 2009 strain was most probably introduced accidentally through the shipping of tomato seedlings and/or horticultural plants (including those serving as reservoir species-mainly of the Solanaceae and Fabaceae families) or viruliferous vector-carrying vegetables for agricultural research purposes from Reunion Island (a French-governed island located approximately 226 km from
Mauritius in the Indian Ocean). Reunion Island might have acquired the virus due to agricultural imports from France, which in turn, might have imported the virus from surrounding regions like Spain, whose fields (like those in Axarquia and Murcia) are highly TYLCV-affected. Although the exact migration route of the ancestral TYLCV lineage cannot be precisely determined, the extent of genetic relatedness between isolates found in different countries, as revealed by phylogenetic analyses, is a good indication of possible migration routes from geographic regions where particular strains were first identified. These possible migration routes should be considered when designing surveillance and phytosanitary measures in order to better control the spread of the disease on the island, especially since there have been reports of TYLCV seed transmission and it is possible that tomato seeds, which are the most common trading commodity for tomato across the globe, are acting as an important undetected source of inoculum. Although seed transmission of TYLCV has not been extensively studied, some authors have reported on the potential transmission of the virus through seeds17.
Although, mutation and recombination are common phenomena in viral genome evolution, one of the factors that greatly contribute to a high rate of mutation in plant viral genomes is the persistent and continued use of susceptible plant varieties which provides breeding ground for the evolution of the virus and eventually favors the emergence of new, possibly more virulent strains. This is the case in Mauritius where the commercial varieties grown in the open field (Swaraksha, Jessica and Typhoon) and in protected cultures (Cetia) are varieties which are susceptible to TYLCD at varying degrees of incidence and severity. A survey among local tomato growers carried out in 2015-2018 revealed that these susceptible varieties continue to remain the preferred varieties grown by tomato producers because of their high market value. Continued cultivation of these susceptible varieties poses a real threat by favoring the ideal host conditions for the replication and mutation of the virus.
CONCLUSION
This study shows that over a period of 9 years, with the continued cultivation of susceptible tomato varieties and the presence of alternate hosts such as Capsicum sp. and Solanum nigrum, which are abundant in almost all geographical regions of the island and which have been both found to act as alternate hosts for TYLCV in Mauritius, the parental isolate underwent multiple mutation events that eventually resulted in possibly novel strains. Continued monitoring of existing TYLCV strains on the island is important to better understand the epidemiology of the virus and devise the best suited control strategies.
SIGNIFICANCE STATEMENT
The TYLCV was first discovered in Mauritius in 2009 and genetically characterized but no further molecular analyses have been conducted since then and the genetic identity of local TYLCV isolates over the past 9 years remains unknown. This study aimed at collecting fresh isolates during the period 2015-2018 and genetically characterizing them to draw a profile of the genetic identity of local TYLCV isolates on the island in order to better understand its epidemiology and be able to advise on appropriate control measures.
REFERENCES
- Takooree, S.D., H. Neetoo, V.M. Ranghoo-Sanmukhiya, S. Hardowar and J.E. van der Waals et al., 2021. First report of black scurf caused by Rhizoctonia solani AG-3 on potato tubers in Mauritius. Plant Dis., 105: 213-213.
- Czosnek, H. and M. Ghanim, 2011. Bemisia tabaci-Tomato Yellow Leaf Curl Virus Interaction Causing Worldwide Epidemics. In: The Whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) Interaction with Geminivirus-Infected Host Plants: Bemisia tabaci, Host Plants and Geminiviruses, Thompson, W.M.O. (Ed.), Springer, Dordrecht, Netherlands, ISBN: 978-94-007-1524-0, pp: 51-67.
- Hanley-Bowdoin, L., E.R. Bejarano, D. Robertson and S. Mansoor, 2013. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol., 11: 777-788.
- Díaz-Pendón, J.A., S. Sánchez-Campos, I.M. Fortes and E. Moriones, 2019. Tomato yellow leaf curl Sardinia virus, a Begomovirus species evolving by mutation and recombination: A challenge for virus control. Viruses, 11.
- Lefeuvre, P., D.P. Martin, G. Harkins, P. Lemey and A.J.A. Gray et al., 2010. The spread of tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog., 6.
- Yan, Z., A.M.A. Wolters, J. Navas-Castillo and Y. Bai, 2021. The global dimension of tomato yellow leaf curl disease: Current status and breeding perspectives. Microorganisms, 9.
- Lefeuvre, P., M. Hoareau, H. Delatte, B. Reynaud and J.M. Lett, 2007. A multiplex PCR method discriminating between the TYLCV and TYLCV-Mld clades of Tomato yellow leaf curl virus. J. Virol. Methods, 144: 165-168.
- Lobin, K., K.L. Druffel, H.R. Pappu and S.P. Benimadhu, 2010. First report of Tomato yellow leaf curl virus in tomato in Mauritius. Plant Dis., 94: 1261-1261.
- Accotto, G.P., J. Navas-Castillo, E. Noris, E. Moriones and D. Louro, 2000. Typing of tomato yellow leaf curl viruses in Europe. Eur. J. Plant Pathol., 106: 179-186.
- Malekzadeh, S., K. Bananej and A. Vahdat, 2011. Serological and molecular identification of Tomato yellow leaf curl virus in Khuzestan Province of Iran. Phytopathol. Mediterr., 50: 303-309.
- Alfaro-Fernández, A., J.A. Sánchez-Navarro, M. Landeira, M.I. Font, D. Hernández-Llópis and V. Pallás, 2016. Evaluation of PCR and non-radioactive molecular hybridization techniques for the routine diagnosis of tomato leaf curl New Delhi virus, tomato yellow leaf curl virus and tomato yellow leaf curl Sardinia virus. J. Plant Pathol., 98: 245-254.
- Davino, S., M. Davino and G.P. Accotto, 2008. A single-tube PCR assay for detecting viruses and their recombinants that cause tomato yellow leaf curl disease in the Mediterranean basin. J. Virol. Methods, 147: 93-98.
- Pérez-Losada, M., M. Arenas, J.C. Galán, F. Palero and F. González-Candelas, 2015. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. E, 30: 296-307.
- Mabvakure, B., D.P. Martin, S. Kraberger, L. Cloete and S. van Brunschot et al., 2016. Ongoing geographical spread of Tomato yellow leaf curl virus. Virology, 498: 257-264.
- Navas-Castillo, J., E. Fiallo-Olivé and S. Sánchez-Campos, 2011. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol., 49: 219-248.
- Rey, M.E.C., J. Ndunguru, L.C. Berrie, M. Paximadis and S. Berry et al., 2012. Diversity of dicotyledenous-infecting geminiviruses and their associated DNA molecules in Southern Africa, including the South-West Indian Ocean islands. Viruses, 4: 1753-1791.
- Kil, E.J., S. Kim, Y.J. Lee, H.S. Byun and J. Park et al., 2016. Tomato yellow leaf curl virus (TYLCV-IL): A seed-transmissible geminivirus in tomatoes. Sci. Rep., 6.
How to Cite this paper?
APA-7 Style
Lobin,
K.K., Lefort,
B.M., Taleb-Hossenkhan,
N. (2024). Genetic Diversity of Tomato Yellow Leaf Curl Virus (TYLCV) in Mauritius. Asian Journal of Plant Pathology, 18(1), 13-20. https://doi.org/10.3923/ajpp.2024.13.20
ACS Style
Lobin,
K.K.; Lefort,
B.M.; Taleb-Hossenkhan,
N. Genetic Diversity of Tomato Yellow Leaf Curl Virus (TYLCV) in Mauritius. Asian J. Plant Pathol. 2024, 18, 13-20. https://doi.org/10.3923/ajpp.2024.13.20
AMA Style
Lobin
KK, Lefort
BM, Taleb-Hossenkhan
N. Genetic Diversity of Tomato Yellow Leaf Curl Virus (TYLCV) in Mauritius. Asian Journal of Plant Pathology. 2024; 18(1): 13-20. https://doi.org/10.3923/ajpp.2024.13.20
Chicago/Turabian Style
Lobin, Kanta, Kumar, Beatrice Melanie Lefort, and Nawsheen Taleb-Hossenkhan.
2024. "Genetic Diversity of Tomato Yellow Leaf Curl Virus (TYLCV) in Mauritius" Asian Journal of Plant Pathology 18, no. 1: 13-20. https://doi.org/10.3923/ajpp.2024.13.20

This work is licensed under a Creative Commons Attribution 4.0 International License.




